DNA transcription: looping genes
18 Feb 2022
LMU scientists have shown that a fundamental process of life works differently than previously assumed.
18 Feb 2022
LMU scientists have shown that a fundamental process of life works differently than previously assumed.
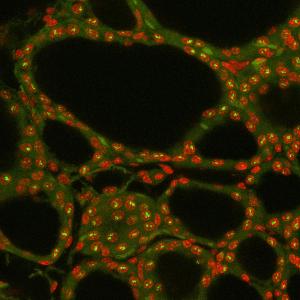
(Nuclei in red, RNA signal in green). Author: I. Solovei
The basic process of life, known as transcription, is the process by which the genetic information stored in DNA is copied by specific enzymes called RNA polymerases into RNA molecules. Despite a huge progress in our understanding of the molecular mechanisms of transcription, so far very little is known about the spatial organization of individual expressed genes. Practically all what we know is that transcription occurs in the nuclear interior, where the most actively transcribed genes are localized. Research carried out by a team led by Irina Solovei at LMU’s Biozentrum, in collaboration with Leonid Mirny (MIT, Cambridge, USA) and others, has now investigated the spatial organization of transcription and the behavior of transcribed genes and discovered a mechanism that contradicts a prevalent concept.
According to the most popular hypothesis, the RNA polymerases are grouped together in a sort of stationary transcription factory, which activated genes approach and pass through. It is known, however, that in gigantic specialized chromosomes the transcription start and termination sites are fixed in space and the expressed sequences loop out. Therefore, in this case, genes remain stationary but RNA-polymerases move along genes
“I always suspected that there could not be two fundamentally different mechanisms of transcription in eukaryotes. This raises the question as to what moves and what remains stationary,” says Solovei. The biologist assumes that the resolution of modern light microscopes is not good enough in most cases to render the transcription loops observed in the giant chromosomes visible. It so happens that, in eukaryotes, frequently expressed genes are often very short – too short for the available resolution – whereas large genes are generally not highly expressed and are therefore unsuitable for transcription studies.
Solovei’s team identified several long and highly expressed genes in mouse cells, which could be resolved under a light microscope, and investigated their structure. The scientists found that these genes unfold during transcription, separating their ends and strongly expanding into the nuclear space. In this way, they form transcription loops which are similar to those of the giant chromosomes.
As the researchers demonstrated, the RNA polymerases move along these loops and carry nascent RNA molecules, undergoing a process of RNA maturation characteristic for eukaryotes. “The shape and expansion of transcription loops from harboring chromosomes suggest that transcription loops are stiff structures,” says Solovei. The authors hypothesize that highly expressed genes become stiff because they are densely decorated with multiple nascent RNAs, which in combination with RNA-binding proteins form voluminous particles.
This hypothesis is supported by a polymer modeling of expressed genes carried out by the group of Leonid Mirny. By simulating the behavior of transcribed genes, the researchers showed that modeled genes recapitulate biological observables only upon acquiring a certain stiffness. “The most striking thing for me is the sheer size of transcription loops – up to 10 micrometers”, says Mirny. “It seems that a single long gene can unfold and traverse most of the nucleus.”
“The results of the study show”, says Heinrich Leonhardt, head of the laboratory Human Biology and BioImaging, “that even in our time dominated by molecular biology, microscopy remains an important and powerful tool to study basic biological questions.”
Susanne Leidescher, Johannes Ribisel, Simon Ullrich, Yana Feodorova, Erica Hildebrand, Alexandra Galitsyna, Sebastian Bultmann, Stephanie Link, Katharina Thanisch, Christopher Mulholland, Job Dekker, Heinrich Leonhardt, Leonid Mirny, Irina Solovei: Spatial organization of transcribed eukaryotic genes. Nature Cell Biology 2022